Developmental Expression of Cytochrome P450 Aromatase Genes (CYP19a and CYP19b) in Zebrafish Fry (Danio rerio)
JOHN M. TRANT¹*, SONJA GAVASSO¹, JARED ACKERS¹, BON-CHU CHUNG², AND ALLEN R. PLACE¹*
¹Center of Marine Biotechnology, University of Maryland Biotechnology
Institute, Baltimore, Maryland 21202
²Institute of Molecular Biology, Academia Sinica, Taiwan
JOURNAL OF EXPERIMENTAL ZOOLOGY 290:475–483 (2001)
*Correspondence to: John M. Trant or Allen R. Place, Center of Marine Biotechnology, University of Maryland Biotechnology Insti- tute, 701 E. Pratt St., Baltimore, MD 21202. E-mail: trant@umbi.umd.edu or place@umbi.umd.edu
Received 19 September 2000; Accepted 30 April 2001
ABSTRACT
Cytochrome P450 aromatase (CYP19) is the terminal enzyme in the steroidogenic pathway that converts androgens (e.g., testosterone) into estrogens (e.g., estradiol). Regulation of this gene dictates the ratio of androgens to estrogens; therefore, appropriate expression of this enzyme is critical for reproduction as well as being pivotal in sex differentiation for most vertebrates. It is assumed that most vertebrates have a single CYP19 gene that is regulated by multiple tissue-specific promoter regions. However, the zebrafish (Danio rerio) has two genes (CYP19a and CYP19b), each encoding a significantly different protein and possessing its own regulatory mechanism. The primary purpose of this study was to determine the pattern of expression of each of the CYP19 genes in the developing zebrafish. A fluorescent-based method of real-time, quantitative RT-PCR provided the sensitivity and specificity to determine transcript abundance in single embryos/juveniles harvested at days 0 through 41 days post-fertilization (dpf), which encompasses the developmental events of sex determination and gonadal differentiation. CYP19 transcripts could be detected as early as 3 or 4 dpf, (CYP19a and CYP19b, respectively) and peak abundance was detected on day five. In general, the CYP19 genes differed significantly in the ontogeny of their expression. In most cases, the gonadal form of CYP19 (CYP19a) was more abundant than the brain form (CYP19b); however, unlike CYP19a, the pattern of CYP19b expression could be clearly segregated into two populations, suggesting an association with sex differentiation. Pharmacological steroids (ethinylestradiol and 17α-methyltestosterone) enhanced the expression of the CYP19b gene at all three days examined (4, 6, and 10 dpf). These data suggest that the timely and appropriate expression of CYP19 is important in development and that the expression of CYP19b (the "extra-gonadal" form) may be associated with sexual differentiation if not sexual determination. J. Exp. Zool. 290:475–483, 2001. © 2001 Wiley-Liss, Inc.
Sex steroids can influence phenotypic sex in all vertebrates but this is especially true for lower vertebrates where complete sex reversal can be induced irrespective of the genetic background (Baroiller et al., ’99). Developing zebrafish can readily be masculinized with 15 days of exposure to 17α-methyltestosterone (Westerfield, ’95). Since exogenous steroids can have such a remarkable influence on phenotypic sex, it is presumed that endogenous sex steroids are key agents in sex differentiation in the developing fry. It is known that the lipophilic nature of yolk retains maternal steroids, however it is unlikely that the uncontrolled release of these endocrine agents during yolk resorption would dictate the sex phenotype. Instead, it is presumed that precise regulation of steroid exposure is required for this important developmental process and the primary mechanism for the control of steroid hormone synthesis is through the transcriptional regulation of the genes that encode steroidogenic enzymes. The terminal enzyme in the steroidogenic pathway is cytochrome P450 aromatase (CYP19; P450arom) and is responsible for the conversion of androgens to estrogens (Simpson et al., ’94). The estrogen, estradiol-17β (E₂), is a sex steroid that functions in many physiological processes including embryo development, onset of puberty, brain sex differentiation, gametogenesis, development of secondary sex characters, behavior, and immune response. Estradiol effects are often in opposition to androgens (e.g., testosterone; T).
Aromatase activity has been found in all vertebrates (Callard, ’83), including the most primitive fishes (hagfish, Kime et al., ’80, and lamprey, Bolduc and Sower, ’92; Kime and Callard, ’82; Sower et al., ’85) and in an invertebrate chordate (amphioxus, Callard et al., ’84). P450arom is expressed in the gonad (typically considered the primary source of the plasma sex steroid hormone in females, Simpson et al., ’94) and in the brain (primarily for local synthesis and action) in all adult vertebrates (Lephart, ’96). In mammals, P450arom is also expressed in a wide variety of “nonsteroidogenic” tissues including placenta, carcinoma, bone, skin, adipose, and breast tissues. The expression of P450arom in lower vertebrates is typically restricted to the gonad, neural tissues (brain and retina), and the pituitary (Callard, ’83; Olivereau and Callard, ’85; Callard et al., ’81a, ’81b; Young et al., ’83; Callard et al., ’93; Trant et al., ’97; reviewed by Lin et al., ’91). To date, the P450arom gene or cDNA has been isolated from a number of fishes (catfish, Trant, ’94; medaka, Tanaka et al., ’95; Fukada et al., ’96; tilapia, Chang et al., ’97; goldfish, Tchoudakova and Callard, ’98; Gelinas et al., ’98; trout, Tanaka et al., ’92; Atlantic stingray, Ijiri et al., 2000), including the zebrafish (CYP19a, GenBank Accession number AF004521; CYP19b, Chung, 2000; Chaing et al., 2001).
In mammals, with the notable exception of the pig (Corbin et al., ’95, ’99; Choi et al., ’97; Conley et al., ’97), there is a single P450arom gene in the genome with multiple promoters (Toda and Shizuta, ’94; Young and McPhaul, ’97; Toda et al., ’90; Means et al., ’91; Zhao et al., ’95; Michael et al. ’95, ’97; Shozu et al., ’97, ’98). However, the goldfish (Carassius auratus; Tchoudakova and Callard, ’98; Gelinas et al., ’98), tilapia, and zebrafish (Danio reria) diverge from the typical vertebrate strategy in that they possess multiple P450arom genes, each of which are regulated differently and encode proteins with different enzymology characteristics. The only other CYP19 genes described in fish are from the stingray (28 kb; Ijiri and Trant, submitted) and medaka (3 kb; Tanaka et al., ’95). There is no information about the multiplicity of CYP19 genes in the medaka but the single stingray CYP19 gene is clearly organized similarly to the mammalian gene.
The purpose of the present study was to examine the developmental changes in the expression of both types of P450arom genes in the young zebrafish as its bipotential gonad differentiates into an ovary or testis. Changes in transcript abundance were determined simultaneously for both of the CYP19 genes and β-actin in single embryo/fry using a highly specific, highly sensitive fluorescent method of real-time quantitative RT-PCR analysis. These data suggest that the “brain form” of CYP19 (CYP19b), as opposed to the “gonadal form” (CYP19a) may be associated with sex determination and differentiation.
MATERIALS AND METHODS
Reagents and supplies
Ethinylestradiol and 17α-methyltestosterone were obtained from Sigma Chemocal Co. (St. Louis, MO). Trizol Reagent and MMLV-reverse transcriptase were purchased from Life Technologies (Gaithersburg, MD). Homogenization tubes for RNA purification (FastPrep Green) were obtained from Bio101 (La Jolla, CA). The TaqMan Universal PCR Master Mix reagent kits and disposable PCR supplies were purchased from PE Biosystems (Foster City, CA).
All oligonucleotide primers were synthesized by the BioAnalytical Services Laboratory at the Center of Marine Biotechnology. Reverse transcription reactions were primed with a “clamped oligo(dT)” with the sequence of 5′-T(16)(ACG)N-3′. Probes and primers for the TaqMan procedure were designed with the assistance of Primer Design (version 1.0) software (PE Biosystems) using the parameters set forth by the TaqMan Sequence Detection system (see below). Fluorescent probes were synthesized by PE Biosystems. TaqMan primer sets were selected so that the amplified cDNA contained a putative exon/exon splice site to which the fluorescent probe hybridized. The primer/probe set for CYP19a was 5′-AGATGTCGAGTTAAAGATCCT- GCA-3′(Forward), 5′-CGACCGGGTGAAAACGT- AGA-3′(Reverse), FAM-5′-ACAGTGTTTTAGCT- GGCCAGAGCCTCCA-3′-TAMRA (Probe). The primer/probe set for CYP19b was 5′-GACAC- TCTCTCCATCAGTCTGTTCTT-3′ (Forward), 5′- CATTCAGTTTCTGCAAGTCAGCA-3′ (Reverse), FAM-5′-CTCGCGAGCCTATCTGAGACTGTATCT- CCTG-3′-TAMRA(Probe). β-actin was used as an internal control to normalize abundance. The actin primer/probe set was 5′-AGGTCATCACCAT- CGGCAAT-3′(Forward), 5′-GATGTCCACGTC- GCACTTCAT-3′ (Reverse), FAM-5′-CTTCCAGCC- TTCCTTCCTGGGTATGGAA-3′-TAMRA(Probe). Sets of forward and reverse oligonucleotide primers used for the determination of CYP gene chromosomal location were 5′-GCTGCAGATCCTG- GAGAGTTTTA-3′ (Forward) and 5′-ATGTTTG- TTCCTTTCTTCACGTTGTA-3′(Reverse), exon 9 of CYP19a), 5′-GCTGACTTGCAGAAACTGAATGTT- 3′(Forward) and 5′-CCAGAGACTGCCTCATGAT- GAA-3′(Reverse), exon 9 of CYP19b), 5′-TCGAGA- TGGGAAAGATATAGCGTAT-3′ (Forward) and 5′- CTGAACCTTCTCCAAACATGCA-3′(Reverse), exon II of CYP17), and 5′-AGACGGTGCTATCCT- GTTCAAAG-3′(Forward) and 5′-CGCCATATTTT- TGTTCCTGTAG-3′(Reverse), exon II of CYP11A).
Animals
Adult zebrafish were maintained at 28°C in recirculating 40 liter aquaria and bred according to established guidelines (Westerfield, ’95). Harvested eggs were immediately transferred to water containing 60 mg sea salts and 0.4 mg methylene blue per liter of pure water. Fry were initially fed a diet of paramecium and then weaned to brine shrimp napulii.
Five to 18 fish were taken at approximately 2-day intervals as indicated in the pertinent figures. All fish were immediately flash frozen on dry ice or in liquid nitrogen and then stored individually in a FastPrep tube at –80°C until analyzed for transcript abundance.
Real-time quantitative RT-PCR analysis (TaqMan)
Whole embryos/fry/juveniles were processed for 20 sec (FastPrep homogenizer, Savant Instruments, Farmingdale, NY) individually in FastPrep Green tubes (Bio101, Vista, CA) with 1 ml of Trizol Reagent supplemented with 4 μl of polyacryl carrier (MRC, Cincinnati, OH). The entire quantity of total RNA (for juveniles up to 30 days post- fertilization (dpf) was used as a template in a 30 μl reverse transcription (RT) reaction using a clamped oligo(dT) primer according to the manufacturer’s recommendation. For each fish, quantitative PCR reactions were performed to determine transcript abundance for each of the three genes. Specifically, duplicate 25 μl reactions containing 300 nM of each TaqMan primer and 100 nM fluorescent probe were cycled in an ABI Model 7700 DNA Sequence Detector (PE Biosystems, Foster City, CA) using the manufacturer’s universal thermal cycling conditions for each transcript. For each set of PCR analyses, all three transcripts were determined in a “standard sample” to determine interassay variability. The computations and descriptions of a modification of this procedure (TaqMan technology) have been published elsewhere (Heid et al., ’96; PE Biosystems User Bulletin, ’97). Transcript abundance of the steroidogenic genes were normalized to the abundance of β-actin and reported as a fold change in abundance relative to the values obtained from a set of animals denoted in the figures.
In one set of analyses, the transcript abundance of CYP19 was determined in brain, liver, and go- nadal tissues dissected from reproductively active adult zebrafish. Total RNA was isolated from 100 mg (or less) of each tissue and quantified by UV260 absorbance. An aliquot of the RNA (1 μg) was reversed transcribed as described above and 1/1000th of this reaction was used in triplicate quantitative fluorescent PCR reactions to determine transcript abundance for each of the three genes.
Mapping the CYP19 genes
PCR analyses with gene-specific primers were used to identify the linkage group of the two CYP19 genes, as well as CYP11A and CYP17 in radiation hybrid (RH) libraries. Two RH libraries are available for mapping—zebrafish genes and markers. The Goodfellow panel (also named RG panel, Goodfellow T51, Zebrafish/Hamster panel, Kwok et al., ’98, ’99 ) was made by fusing irradiated zebrafish AB9 cells with Hamster Wg3H cells. The Ekker panel (also named LN54 panel, Ekker et al., ’96) was made by fusing irradiated AB9 cells with Mouse B78 cells. The potential resolution of the Goodfellow panel and the Ekker panel are 350 KB and 500 KB, respectively.
The RH panel was scored according to Hudson et al. (’95). Caution is required when scoring the hybrids, since the donor chromosomal fragments are present at various molarities among the hybrid cell lines; therefore, all assays were performed at least twice. Discordance between the multiple runs of a marker was at or below 5%. The results indicate in which linkage group (LG) the gene is located and the centiRay distance (cR; defined as a percent frequency of a breakage occurring between two markers after exposure to a specific radiation dose) between markers. For the LN54 panel, 1cR is approximately equal to 148 kilobases. Full maps for the LGs containing the closest marker can be found at ZFIN (http://zfin.org/ ZFIN/). “Z” markers are from the MGH meiotic map and cDNAs can be found on the MOP meiotic map at the same web site. To be considered significant, the best lod score (logarithm of the likelihood ratio for linkage) must be greater than a lod of five, and the net difference between the best lod score and the lod score on the second best LG should be at least a difference of three lods.
RESULTS
In this study, a highly sensitive and specific quantitative RT-PCR method (TaqMan) was developed to simultaneously detect transcript abundance of both forms of CYP19 and β-actin in adult tissues and individual fry/juvenile zebrafish. The assay was characterized to ensure linearity of response, specificity of the assay, and lack of interference from contaminating genomic DNA (up to 50 ng of genomic DNA). The coefficient of interassay variability was 6.04%.
CYP19 expression in adults
The abundance of transcripts derived from the CYP19a and CYP19b genes expressed in the brain, gonads, and liver (negative control) of adult male and female zebrafish indicates that both forms were expressed in both the gonads and the brains (Fig. 1). Expression of both genes was relatively equivalent in the testis; there was about a 300 fold difference in transcript abundance in the brain (where CYP19b predominated) and the ovary (where CYP19a predominated). In fact, the level of expression of CYP19b was higher in the brains of females than was the expression of CYP19a in the ovary. This is contrary to the typical pattern of tissue-specific expression since the ovary is generally considered the estrogenic organ of vertebrates. Even though the abundance of both forms of the P450arom transcripts was similar and consistent, the abundance of CYP19b transcript in the brain could clearly be divided into two categories (n = 3 and 5 for low and high expression, respectively). Differences in the physiology of these two groups of males have yet to be identified. All males were reproductive adults at the time of sacrifice but the age of the individuals and/or the behavioral status or rank of males was not known.


CYP19 expression in embryos and fry
As conducted in the above study for adults, the TaqMan quantitative PCR method was used to simultaneously determine the transcript abundance of CYP19a and CYP19b (in addition to β- actin) in whole individual embryos/fry. Individuals were taken from 0 to 41 dpf, which encompasses the developmental events of hatching (2.5 dpf), sex determination, and gonadal differentiation. It has been shown that primordial ovarian follicles can be seen by histological examination as early as day 21 and the onset of testicular development lags behind until day 25 to 28 (Chiang et al., 2001).
CYP19 transcripts were not detected until after hatching. However it is clear that the expression of the CYP19b gene (Fig. 2) and the CYP19a gene (Fig. 3) were developmentally regulated in the growing fry prior to the development of the bipotential gonad into a testis or ovary. To ensure that this pattern of CYP19 expression was not merely a reflection of actin (to which the CYP19 data were normalized), the changes in abundance of the actin transcript in the entire animal were examined. Fig. 4 clearly indicates a relatively steady increase in actin transcript abundance up to 63 fold (expressed relative to the actin abun- dance detected in 24 and 48 hr embryos) through day 16. By day 18, actin transcript abundance increases several thousand fold, thereby resulting in an underestimation of the abundance of the CYP19 transcripts. However, such dramatic changes are not reflected at this time period in CYP19 expression shown in Figs. 2 and 3. Although the actin data was not mathematically normalized, procedurally, these data reflect the changes in “whole body” actin transcript abundance since differences in the recovery of RNA was minimized by using a co-precipitate, and the sta-tistical efficiencies of the RT and PCR reactions are designed to be unified.
CYP19a and CYP19b transcripts were detected as early as 3 or 4 dpf, respectively. The expression of CYP19b was dynamically variable throughout development and growth of the fry. The pattern of CYP19b expression was clearly segregated into two populations except for three short periods (0 to 4 days, 9 to 11 days, and 19 to 22 days). All of the “low means” from days 0 to 16 were set to zero since CYP19b transcript was not detectable in these animals. For clarity, the differences in the two populations are highlighted in Fig. 2 by lines that merely connect all of the high mean values and all of the low mean values. Although it is assumed these categorical differences in the means of transcript abundance were sex-linked, there is no data to suggest that one pattern of expression was associated with a particular sex. It is interesting that the CYP19b transcript could not be detected from day 9 to day 11, and that one group or the other does not express CYP19b either before (6–8 days) or after (13–16 days) this period.
The highest abundance of CYP19a transcript was detected immediately after hatching (Fig. 3). More importantly, there appears to be a single, regulated pattern of CYP19a expression in the same fish that displayed two patterns of CYP19b expression. However, using whole fry for these analyses is misleading since the tissue-specific expression of these genes may be the critical parameter. In order to partially address this concern, a preliminary experiment was conducted where fry were dissected into “cephalic” and “trunk” sections. TaqMan analyses found that the CYP19a transcript was expressed in both sections of the fry whereas the CYP19b form was expressed within the cephalic section only (results not shown).
Disruption of CYP19 expression
A preliminary experiment was conducted where zebrafish fry were taken at 4, 6, and 10 dpf after continuous exposure to 170 nM ethinylestradiol (EE; a pharmaceutical estrogen) or 17α-methylt-estosterone (MT; an aromatizable pharmaceutical androgen routinely used for masculinization of a number of species of fish, including zebrafish). The data indicate that the expression of the CYP19b gene was dramatically altered (Fig. 5), whereas the expression of the CYP19a gene was essentially unaffected (not shown). Exposure to either pharmaceutical steroid induced the expression of the CYP19b gene approximately four fold higher than the controls and expression was maintained on day ten when CYP19b expression was not detectable in the controls. Presumably, the differential effect of the steroids on the expression of the two genes was due to an estrogen-responsive cis element (ERE) in the 5′-flanking region of the CYP19b gene but not the CYP19a gene (manuscript in preparation). The differential responsiveness of the genes to estrogens is consistent with our description of the Atlantic stingray CYP19 gene (Trant and Ijiri, 2000) and with the description of the zebrafish genes from the laboratory of Dr. Gloria Callard (presented at the 4th International Symposium on Fish Endocrinology, Seattle, WA). Even though the zebrafish (and probably all other teleosts) have utilized a strategy of gene duplication to handle the need for differential and tissue-specific expression of P450arom, the promoter elements are likely to be very similar to that seen in the CYP19 gene of other vertebrates.
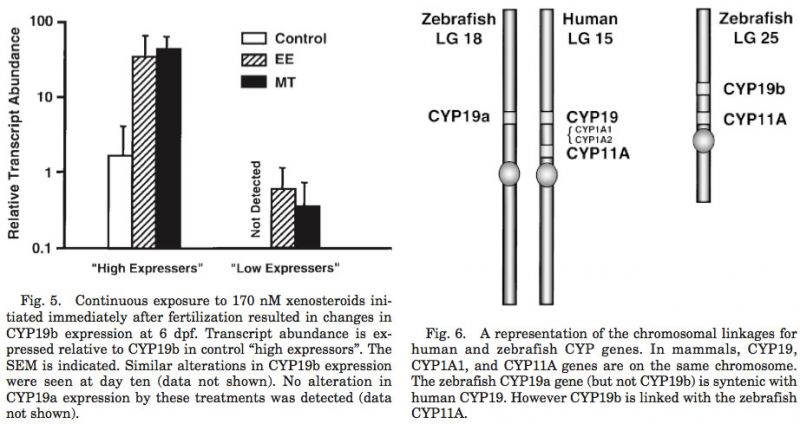
Synteny of the CYP19 genes
CYP19a mapped to linkage group 18 (LOD = 15.5; 6.08 cR from Z8343 in LN54), which is syntenic to linkage groups 15 and 9 of the human and mouse, respectively (Fig. 6). CYP19b mapped to the linkage group 25 (LOD = 10.0; 15.55 cR from Z20832 in LN54). Interestingly, the CYP11A gene that is linked to CYP19 in mammals (i.e., LG 18 of man) mapped with the CYP19b gene on LG 25, as opposed to the expected link to the CYP19a gene (LOD = 20.37; 31 cR from fb24b05 in T51). The zebrafish CYP17 gene (LG 13; LOD = 15.33; 7 cR from z13250_13 in T51) was found to be syntenic to the mammalian CYP17 gene (linkage groups 10 and 19 in man and mouse, re- spectively). Only a single copy of the CYP11A and the CYP17 genes has been identified in the zebrafish to date. Therefore, these data will need to be re-evaluated if it is determined that these genes were duplicated in the zebrafish genome.
DISCUSSION
It is becoming increasingly evident that teleosts possess two forms of the aromatase genes (CYP19a and CYP19b) in their genome, and the cDNA encoding both forms have been isolated from the zebrafish (Chaing et al., 2001). As shown in this report (Fig. 1) and originally shown in the goldfish (Tchoudakova and Callard, ’98), both genes are expressed in gonads and in estrogenic extra-gonadal tissues (such as brain and pituitary). However, the transcript for CYP19a is 100 fold more abundant in the adult ovary whereas CYP19b predominates in extra-gonadal tissues. Both forms of the CYP19 gene are equally expressed in the zebrafish testis. Although we have no explanation, it is interesting that CYP19b expression in the male brain is quite variable and our data may represent two physiological and/or behavioral states.
The highly sensitive and specific TaqMan method of real-time, quantitative fluorescent RT-PCR allows for the simultaneous detection of transcripts from multiple genes in extremely small pieces of tissues, such as a single zebrafish embryo/fry. Using this method, it was determined that CYP19 is expressed as early as 3 dpf in the developing zebrafish fry/juvenile, which is well before ovarian (21 dpf) or testicular (25–28 dpf) differentiation can be determined by histological examination. Both CYP19a and CYP19b are expressed the highest almost immediately after hatching (3 or 4 dpf, respectively); thereafter, however, the two genes have substantially different patterns of expression. These patterns of CYP19 expression were not influenced by the potential changes in actin expression (used for normalization of the data) during development and growth of the fry/juvenile (Fig. 4). Most importantly, the changes in actin expression during the first two weeks were rather constant, during which time CYP19 transcripts varied dramatically. It is clear that the differential regulation of the two CYP19 genes was developmentally regulated and this conclusion is reflected in the differences in the promoter elements identifiable in their respective 5′-flanking regions of the gene (unpublished data).
It is interesting that CYP19a is expressed in greater abundance than CYP19b; however, it appears that CYP19b expression is sexually dimorphic. It should be noted that the lines connecting the data points of CYP19b expression (Fig. 2) are purely arbitrary. However, it is clear that there are two distinct groups at most time points examined in this study. This dichotomy was not detected with CYP19a; therefore, our hypothesis is that it is the expression of the CYP19b gene that dictates phenotypic sex differentiation, if not sex determination. Of course, until in situ hybridization experiments identify the site(s) of CYP19b expression in the bipotential fry, it cannot be determined if this estrogen signal arises from the neural tissues (brain, pituitary, spinal cord, or gonadal neurons) or from germ and accessory cells of the primordial gonad. In either case, these data suggest that the developmental regulation of CYP19 via the extra-gonadal promoter may be a critical aspect of sex determination / differentiation that has been poorly investigated. This aspect of sex differentiation becomes critical with respect to the study of environmental xenoestrogens since the extra-gonadal promoter of CYP19 contains estrogen responsive elements (ERE), whereas the gonadal promoter lacks these elements and is unresponsive to estrogens.
It has been well established that exogenous steroids can dictate phenotypic sex independent of the genetic background of the developing fry. In fact, the use of 17α-methyltestosterone (MT) is often used to masculinize zebrafish (Westerfield, ’95). Although attempts have been made, feminization of zebrafish with xenoestrogens has been unsuccessful. In order to address some of the questions of sex differentiation and its disruption by xenosteroids, a study was conducted to examine the changes in CYP19a and CYP19b expression induced by MT during the accepted protocol for sex reversal. For comparison, the effects of a pharmaceutical estrogen (ethinylestrodiol; EE) during this exposure were also examined. As shown before, at 6 dpf there were two populations of means for CYP19b in control fry and this trend was also evident in the fry exposed to xenosteroids. As expected, the xenoestrogen had no effect on CYP19a expression at 6 dpf but it substantially elevated CYP19b transcript abundance (Fig. 5). As with the controls, there were two populations of CYP19 expression in the fry and these data have been arbitrarily segregated into “low expressors” and “high expressors”. Identical results were seen with MT, presumably due to aromatization of MT (17α-methylestradiol) interacting with the ERE of the CYP19b gene in order to induce expression. Although the animals of this experiment were not evaluated for sex reversal, it is presumed that the remaining nonmetabolized MT was of sufficient concentration to induce masculinization. Similar results were seen at day ten when CYP19b expression was undetectable in the controls (data not shown) but readily detectable in the EE- and MT-exposed fry. Since excess estrogens have no effect on phenotypic sex of zebrafish, these data strongly suggest that androgens can overcome the effects of endogenous estrogens. It is unknown if this effect is mediated via the activation of androgen-responsive genes and/or the down regulation of the CYP19 genes. The androgen-mediated regulation of CYP19 expression needs to be examined (see Jeyasuria and Place, ’98), as well as a description of the phenotype of zebrafish fry exposed to inhibitors of both aromatases.
Mapping of the CYP19 genes in the zebrafish genome provides some insights as to the tetraploidization event that gave rise to the teleosts and potential steroidogenic genes that have yet to be isolated. The CYP19a and the CYP17 genes were syntenic with the respective genes of the human and mouse. However, the zebrafish CYP11A (cholesterol side chain cleavage) was not. Since CYP19 and CYP11A are closely linked, these data strongly suggest that there is an additional isoform of CYP11A in the zebrafish genome, and we predict that it will map with CYP19A on the zebrafish LG18. If true, then by extension, one would predict that the xenobiotic metabolizing hepatic enzyme, CYP1A, would have also been duplicated. Only a single form of the cDNA encoding CYP1A has been isolated in the zebrafish, and it has not been mapped. The isolation of a second isoform of CYP1A and/or CYP11A would verify the duplication of large linkage groups of the teleost genome. More importantly, as with the CYP19 genes, the interplay between the two isoforms of these genes will be very important for the study of steroid endocrinology (CYP11A) and toxicology (CYP1A) in fish.
In summary, the “extra-gonadal” form of the P450arom gene (CYP19b) is highly expressed in the adult zebrafish brain with transcript abundances exceeding the gonadal form (CYP19a) in the ovary. In addition, it appears that it is the differential expression of the extragonadal form prior to sex differentiation that dictates the pathway for phenotypic sex, even though the gonadal form of the transcript is in greater abundance in the whole fry.
ACKNOWLEDGMENTS
This research was supported by grants from the Wallenburg Foundation and NSF (IBN-9514051) to J.M.T., and grants from NIEHS (ES09563-03) and NSF (IBN96-04265) to A.R.P. This is contribution #543 from the UMBI Center of Marine Biotechnology.
LITERATURE CITED
Baroiller J–F, Guiguen Y, Fostier A. 1999. Endocrine and environmental aspects of sex differentiation in fish. Cell Mol Life Sci 55:910–931.
Bolduc TG and Sower SA. 1992. Changes in brain gonadotropin-releasing hormone, plasma estradiol-17β, and progesterone during final reproductive cycle of the female sea lamprey, Petromyzon marinus. J Exp Zool 264:55–63.
Callard GV. 1983. Androgen and estrogen actions in the vertebrate brain. Am Zool 23:607–620.
Callard GV, Petro Z, Ryan K. 1981a. Estrogen synthesis in vitro and in vivo in the brain of a marine teleost (Myoxocephalus). Gen Comp Endocrinol 44:359–364.
Callard GV, Petro Z, Ryan K. 1981b. Biochemical evidence for aromatization of androgen to estrogen in the pituitary. Gen Comp Endocrinol 44:364–369.
Callard GV, Pudney J, Kendall SL, Reinboth R. 1984. In vitro conversion of androgen to estrogen in amphioxus gonadal tissues. Gen Comp Endocrinol 56:53–58.
Callard GV, Drygas M, Gelinas D. 1993. Molecular and cellular physiology of aromatase in the brain and retina. J Steroid Biochem Mol Biol 44:541–547.
Chang XT, Kobayashi T, Kajiura H, Nakamura M, Nagahama Y. 1997. Isolation and characterization of the cDNA encoding the tilapia (Oreochromis niloticus) cytochrome P450 aromatase (P450arom): changes in P450arom mRNA, protein and enzyme activity in ovarian follicles during oogenesis. J Mol Endocrinol 18:57–66.
Chiang EF-L, Yan Y-L, Tong S-K, Hsiao P-H, Guiguen Y, Postlethwait J, Chung B-C. 2001. Characterization of duplicated zebrafish cyp 19 genes. Second International Symposium on the Biology of Vertebrate Sex Determination. Honolulu, Hawaii. April 10–14, 2000.
Choi I, Troyer DL, Cornwell DL, Kirby-Dobbels KR, Collante WR, Simmens FA. 1997. Closely related genes encode developmental and tissue isoforms of porcine cytochrome P450 aromatase. DNA Cell Biol 16:769–777.
Chung B-C. 2000. Expression of steroidogenic P450 genes in transgenic mouse and zebrafish. In: Ohamoto M, Ishimura Y, Nawata H, editors. Molecular Steroidogenesis. Tokyo, Japan: Universal Academy Press, Inc. p 317–320.
Conley AJ, Corbin CJ, Smith T, Hinshelwood M, Liu Z, Simpson ER. 1997. Porcine aromatases: studies on tissuespecific, functionally distinct isozymes from a single gene? J Steroid Biochem Mol Biol 61:407–413.
Corbin CJ, Khalil MW, Conley AJ. 1995. Functional ovarian and placental isoforms of porcine aromatase. Mol Cell Endocrinol 113:29–37.
Corbin CJ, Trant JM, Walters KW, Conley AJ. 1999. Changes in testosterone metabolism associated with the evolution of placental and gonadal isozymes of porcine aromatase cytochrome P450. Endocrinology 140:5202–5210.
Ekker M, Speevak MD, Martin CC, Joly L, Giroux G, Chevrette M. 1996. Stable transfer of zebrafish chromosome segments into mouse cells. Genomics 33(1):57–64.
Fukada S, Tanaka M, Matsuyama M, Kobayashi D, Nagahama Y. 1996. Isolation, characterization and expression of cDNAs encoding the medaka (Oryzias latipes) ovarian follicle cytochrome P-450 aromatase. Mol Reprod Dev 45:285–290.
Gelinas D, Pitoc GA, Callard GV. 1998. Isolation of a goldfish brain cytochrome P450 aromatase cDNA: mRNA expression during the seasonal cycle and after steroid treatment. Mol Cell Endocrinol 138:81–93.
Heid CA, Stevens J, Livak KJ, Williams PM. 1996. Real time quantitative PCR. Genome Res 6:986–994.
Hudson TJ, Stein LD, Gerety SS, Ma J, Castle AB, Silva J, Slonim DK, Baptista R, Kruglyak L, Xu SH. 1995. An STS-based map of the human genome. Science 270(5244): 1945– 1954.
Ijiri S, Berard C, Trant JM. 2000. Characterization and expression of the cDNA encoding the Atlantic stingray (Dasyatis sabina) form of cytochrome P450 aromatase (CYP19). Mol Cell Endocrinol 164:169–181.
Jeyasuria P and Place AR. 1998. Embryonic brain-gonadal axis in temperature-dependent sex determination of reptiles: a role for P450 aromatase (CYP19). J Exp Zool 281: 428–449.
Kime D and Callard GV. 1982. Formation of 15 β-hydroxylated androgens by the testis and other tissues of the sea lamprey, Petromyzon marinus, in vitro. Gen Comp Endocrinol 4:267–270.
Kime D, Hews EA, Rafter J. 1980. Steroid biosynthesis by testes of the hagfish Myxine glutinosa. Gen Comp Endocrinol. 41:8–13.
Kwok C, Korn RM, Davis ME, Burt DW, Critcher R, McCarthy L, Paw BH, Zon LI, Goodfellow PN. 1998. Characterization of whole genome radiation hybrid mapping resources for non-mammalian vertebrates. Nucleic Acids Res 26(15):3562–3566.
Kwok C, Critcher R, Schmitt K. 1999. Construction and characterization of zebrafish whole genome radiation hybrids. Methods Cell Biol 60:287–302.
Lephart ED. 1996. A review of brain aromatase cytochrome P450. Brain Res Rev 22:1–26.
Lin Y-WP, Petrino TR, Wallace RA. 1991. A non-salmonid model for the study of fish reproduction. In: Scott AP, Sumpter JP, Kime DE, Rolfe MS, editors. Proc. of Fourth Intern. Symp. on the Reproductive Physiology of Fish. Fish Symp 91. Sheffield, UK. Norwich, UK: University of East Anglia. p 74–76.
Means GD, Kilgore MW, Mahendroo MS, Mendelson CR, Simpson ER. 1991. Tissue specific promoters regulate aromatase cytochrome P450 gene expression in human ovary and fetal tissues. Mol Endocrinol 5:2005–2013.
Michael MD, Kilgore MW, Morohashi K, Simpson ER. 1995. Ad4BP/SF-1 regulates cyclic AMP-induced transcription from the proximal promoter (PII) of the human aromatase P450 (CYP19) gene in the ovary. J Biol Chem 270:13561– 13566.
Michael MD, Michael LF, Simpson ER. 1997. A CRE-like sequence that binds CREB and contributes to cAMP-dependent regulation of the proximal promoter of the human aromatase P450 (CYP19) gene. Mol Cell Endocrinol 134:147–156.
Olivereau M, Callard G. 1985. Distribution of cell types and aromatase activity in the sculpin (Myoxocephalus) pituitary. Gen Comp Endocrinol 58:280–290.
PE Applied Biosystems. 1997. ABI PRISM 7700 Sequence Detection System. User Bulletin #2.
Shozu M, Zhao Y, Simpson ER. 1997. Estrogen biosynthesis in THP1 cells is regulated by promoter switching of the aromatase (CYP19) gene. Endocrinology 138:5125–5135.
Shozu M, Zhao Y, Bulun SE, Simpson ER. 1998. Multiple splicing events involved in regulation of human aromatase expression by a novel promoter, I.6. Endocrinology 139: 1610–1617.
Simpson ER, Mahendroo MS, Means GD, Kilgore MW, Hinshelwood MM, Graham-Lorence S, Amarneh B, Ito Y, Fisher CR, Michael MD, Mendelson CR, Bulun SE. 1994. Aromatase cytochrome P450, the enzyme responsible for estrogen biosynthesis. Endocr Rev 15:342–355.
Sower SA, Plisetskaya E, Gorbman A. 1985. Changes in plasma steroid and thyroid hormones in insulin during final maturation and spawning of the sea lamprey, Petromyzon marinus. Gen Comp Endocrinol 58:259–269.
Tanaka M, Telecky TM, Fukada S, Adachi S, Chen S, Nagahama Y. 1992. Cloning and sequence analysis of the cDNA encoding P-450 aromatase (P450arom) from a rainbow trout (Oncorhynchus mykiss) ovary; relationship between the amount of P450arom and the production of oestradiol-17β in the ovary. J Mol Endocrinol 8:
Tanaka M, Fukada S, Matsuyama M, Nagahama Y. 1995. Structure and promoter analysis of the cytochrome P-450 aromatase gene of the teleost fish, medaka (Oryzias latipes). J Biochem (Tokyo) 117:719–725.
Tchoudakova A, Callard GV. 1998. Identification of multiple CYP19 genes encoding different cytochrome P450 aromatase isozymes in brain and ovary. Endocrinology 139:2179–2189.
Toda K, Shizuta Y. 1994. Identification and characterization of cis-acting regulatory elements for the expression of the human aromatase cytochrome P-450 gene. J Biol Chem 269:8099–8107.
Toda K, Terashima M, Kawamoto T, Sumimoto H, Yokoyama Y, Kuribayashi I, Mitsuuchi Y, Maeda T, Yamamoto Y, Sagara Y, Ikeda H, Shizuta Y. 1990. Structure and functional characterization of human aromatase P-450 gene. Eur J Biochem 193:559–565.
Trant JM. 1994. Isolation and characterization of the cDNA encoding the channel catfish (Ictalurus punctatus) form of cytochrome P450arom. Gen Comp Endocrinol 95:155–168.
Trant JM, Ijiri S. 2000. Molecular studies of steroidogenesis using primitive vertebrates (sharks and rays) as models. In: Okamoto M, Ishimura Y, Nawata H, editors. Molecular Steroidogenesis. Tokyo, Japan: Universal Academy Press, Inc. p 313–316.
Trant JM, Lehrter JM, Gregory T, Nunez BS, Wunder J. 1997. Expression of cytochrome P450 aromatase in the channel catfish, Ictalurus punctatus. J Steroid Biochem Mol Biol 61:393–397.
Westerfield M. 1995. The zebrafish book—a guide for the laboratory use of zebrafish (Danio rerio), 3rd edition. Eugene, Oregon: University of Oregon Press.
Young G, Kagawa H, Nagahama Y. 1983. Evidence for a decrease in aromatase activity in the granulosa cells of amago salmon (Oncorhynchus rhodurus) associated with final oocyte maturation. Biol Reprod 29:310–315.
Young M, McPhaul MJ. 1997. Definition of the elements required for the activity of the rat aromatase promoter in steroidogenic cell lines. J Steroid Biochem Mol Biol 61:341–348.
Zhao Y, Mendelson CR, Simpson CR. 1995. Characterization of the sequences of the human CYP19 (aromatase) gene that mediate regulation by glucocorticoids in adipose stromal cells and fetal hepatocytes. Mol Endocrinol 9: 340–349.
Все поля со звездочкой (*) обязательны для заполнения
Данное действие необратимо.
чтобы сделать работу на сайте еще удобнее!
С помощью личного кабинета Вы сможете:
- моментально получать счета на оформленные заказы;
- отслеживать статусы выполнения заказа по оплате, отгрузке, наличию товаров на складе;
- вести историю заказов, повторять заказы полностью или частично;
- выбирать персонального менеджера;
- формировать списки избранного среди товаров, справочных материалов и видео;
- делать заказ со страницы избранных товаров;
- экономить время при заполнении форм заказа по каталогам и регистрации на мероприятия.

